Berkeley Lab researchers create first of its kind gene map of sulfate-reducing bacterium
Related images
(click to enlarge)
Critical genetic secrets of a bacterium that holds potential for removing toxic and radioactive waste from the environment have been revealed in a study by researchers with the U.S. Department of Energy (DOE)'s Lawrence Berkeley National Laboratory (Berkeley Lab). The researchers have provided the first ever map of the genes that determine how these bacteria interact with their surrounding environment. "Knowing how bacteria respond to environmental changes is crucial to our understanding of how their physiology tracks with consequences that are both good, such as bioremediation, and bad, such as biofouling," says Aindrila Mukhopadhyay, a chemist with Berkeley Lab's Physical Biosciences Division, who led this research. "We have reported the first systematic mapping of the genes in a sulfate-reducing bacterium -- Desulfovibrio vulgaris - that regulate the mechanisms by which the bacteria perceive and respond to environmental signals."
Mukhopadhyay, who also holds an appointment with the Joint BioEnergy Institute (JBEI), a DOE Bioenergy Research Center, is the corresponding author of a paper that describes this research in the journal Genome Biology.
Desulfovibrio vulgaris is an anaerobic bacterium that is present in numerous ecological niches and serves as a model organism for the study of sulfate-reducing bacteria. The microbe has drawn much attention -- both good and bad -- for its unique ability to metabolize metals. On the good side, D. vulgaris can generate enzymes that reduce toxic heavy metals and radioactive nuclides into non-hazardous forms. On the bad side, D. vulgaris is also notorious as a pest that corrodes the metals used in oil drilling and storage operations.
"For all of these reasons, it is important that we understand the molecular signaling systems by which D. vulgaris interacts with and survives in its many different environments," says Mukhopadhyay. "Yet, after more than seven decades of study, not a single one of the approximately 70 known molecular signaling systems in D. vulgaris had been characterized."
As humans, it is customary to think of us interacting with our environment through the five senses -- sight, sound, smell, touch and taste. However, this information processing actually takes place through molecular signaling systems. Bacteria also process signals at the molecular level but they utilize a two-component system in which one protein -- a histidine kinase -- senses an environmental signal, which it then transfers to a second protein -- a response regulator -- that controls the reaction.
"These microbial systems are difficult to identify and study because they don't become active until they sense a specific environmental signal and we don't know what most of those signals are," Mukhopadhyay says. "We had to figure a way around this conundrum."
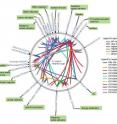
Mukhopadhyay and her co-authors were able to bypass the need to know the signal activation conditions and map virtually the entire D. vulgaris gene response network through genome-wide in vitro experimental determinations. They accomplished this using a "DNA-Affinity-Purified-chip (DAP-chip) strategy" they devised, in which purified response regulator proteins are incubated with genomicDNA and used to enrich DNA regions that bind to them. Both the enriched and the starting input DNA are amplified, pooled and hybridized in a customized D. vulgaris microarray to determine enriched gene targets.
"To our knowledge, this is the first extensive use of a genome-wide method to map all bacterial two component system response regulator binding sites in a single study," Mukhopadhyay says.
Mukhopadhyay and her colleagues have already used their new gene map to predict the functions of several response regulators in D. vulgaris that include key processes of carbon, nitrogen and energy metabolism, cell motility and biofilm formation. They have also predicted responses to stresses such as nitrite, low potassium and phosphate starvation.
"For several response regulators we predicted and experimentally verified the binding site motifs, most of which were discovered as part of this study," Mukhopadhyay says. "In the future this gene map should help guide the development of bioremediation methods that do not exacerbate existing problems, and also help guide field practices that will enhance desirable outcomes."
The DAP-chip strategy used to create this regulatory gene map for D. vulgaris can also be used to create similar gene maps for any microbe whose genome has been sequenced. Given that the regulatory network of a microbe is often a reflection of the environments in which it thrives and the biogeochemical processes it can mediate, such gene maps should have an important role in future clean-ups of a wide range of contaminated environments.
"Our study is inherently foundational and the regulatory networks we discovered not only inform us about the bacterial response to heavy metals but also allows us to correlate microbial activity to other biofouling phenomena as well," says Mukhopadhyay.
Co-authoring the Genome Biology paper on this work, which was carried out through Berkeley Lab's ENIGMA program, were Lara Rajeev, Eric Luning, Paramvir Dehal, Morgan Price and Adam Arkin.
This research was funded by the DOE Office of Science.
Source: DOE/Lawrence Berkeley National Laboratory
Other sources
- First of its kind gene map of sulfate-reducing bacterium createdfrom Science DailyWed, 9 Nov 2011, 19:30:43 UTC
- Researchers create first of its kind gene map of sulfate-reducing bacteriumfrom PhysorgWed, 9 Nov 2011, 18:00:57 UTC