Stanford team develops single cell genomics technique to reverse engineer developing lung
Consider the marvel of the embryo. It begins as a glob of identical cells that change shape and function as they multiply to become the cells of our lungs, muscles, nerves and all the other specialized tissues of the body. Now, in a feat of reverse tissue engineering, Stanford researchers have begun to unravel the complex genetic coding that allows embryonic cells to proliferate and transform into all of the specialized cells that perform a myriad of different biological tasks.
A team of interdisciplinary researchers took lung cells from the embryos of mice, choosing samples at different points in the creature's development cycle. Using the new technique of single-cell genomic analysis, they recorded what genes were active in each cell at each point. Though they studied lung cells, their technique is applicable to any type of cell.
"This lays out a playbook for how to do reverse tissue engineering," said Stephen Quake, the Lee Otterson Professor in the School of Engineering and leader of the research team.
He detailed the experiments in a Nature paper co-authored with Mark Krasnow, a professor of biochemistry at the Stanford School of Medicine and Tushar Desai, an assistant professor of pulmonary and critical care medicine at Stanford.
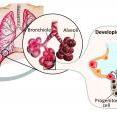
They used this reverse engineering approach to study the cells in the alveoli -- the small balloon-like structures at the tips of the airways. The alveoli serve as docking stations where blood vessels receive oxygen and deliver carbon dioxide.
Barbara Treutlein, a postdoctoral scholar in Quake's lab, together with Doug Brownfield, a postdoctoral researcher in Krasnow lab, isolated 198 lung cells from mouse embryos at three stages of gestation: 14.5 days, 16.5 days and 18.5 days (mice are on average born at 20 days). They also took some lung cells from adult mice.
They used standard enzymatic techniques to dissolve the proteins that hold the lung cells together in tissue form, then sorted out the specific alveolar cell types that were the focus of their study.
Their next steps involved newer techniques at the heart of their reverse engineering process.
Recall how eyedroppers work. Squeeze the rubber bulb to evacuate the air; plunge it into a solution to fill it with fluid; squeeze the bulb again to force the fluid out. In recent years biotechnologists have used those basic principles to develop microfluidic devices of such precision that they can suck a single cell out of solution and isolate it in a chamber to study its genetic material.
Quake's lab has pioneered the use of microfluidic chips to study single cells. In this study, they used microfluidic devices to capture their 198 sample lung cells. Then they used single cell genomic sequencing to measure which genes were active in each cell at each time.
How did they decode genomic activity in a single cell? Recall that DNA in the nucleus of every cell contains the full genome for that organism. That's why it's possible to build an organism from a single cell. But only some of those genes are active in any given cell at any given time. That's why lung cells are different than hair cells; each cell has a different set of active genes directing its functions.
Genes direct cellular activity by making or "expressing" a messenger RNA or mRNA. Each mRNA instructs the cell to make a particular protein. Cells are essentially a group of interacting proteins. Therefore knowing which mRNAs are active offers a lens into the function of that cell at the point when it was captured in the microfluidic device.
Using this process the Stanford researchers revealed for the first time precisely which genes regulate the development of these particular lung cells at each step along the way to mature alveoli.
One important finding involved the development of two important cells types at the tip of the alveoli, where the lung meets blood to perform the gas exchange that keeps us alive.
Alveolar type I cells are the flattest cells in the body. Blood cells dock alongside them to deliver oxygen or pick up carbon dioxide. The thinness of the cell is vital to facilitating this gas transfer.
Alveolar type II cells are compact and cuboidal. They secrete proteins to keep the alveoli from collapsing like empty balloons, so as to maintain the inner space through which oxygen and carbon dioxide can move.
Using single cell genomics allowed the researchers to reverse engineer the development process to show how a single progenitor cell type gives rise to both of these different, mature alveolar cells.
The researchers also captured cells in transition from the progenitor to mature cell state, gaining crucial insights into the mechanism of alveolar cell differentiation
Although this study focused on lung cells, the technique -- capturing individual cells at different stages of embryonic development and assessing gene activity through mRNA sequencing -- can be used to reverse-engineer other tissues.
In addition to studying embryonic development, the technique could be used in clinical settings. For instance researchers could study differences between individual cells in a tumor, improving our understanding of the stages of cancers and leading to better, more targeted therapies.
"This technology represents a quantal leap forward in our ability to apprehend the full diversity of cell types in a given population, including rare ones that could have special functions," said Desai. "Because a comprehensive molecular characterization of each type is achieved, including the signals they send and receive, a snapshot of the communication between individual cells will also emerge and may suggest attractive therapeutic targets in disease."
Source: Stanford School of Engineering
Other sources
- Scientists develop ‘playbook’ for reverse engineering tissuefrom Science BlogThu, 17 Apr 2014, 0:10:19 UTC
- Single cell genomics technique developed to reverse-engineer developing lungfrom Science DailyTue, 15 Apr 2014, 7:19:34 UTC
- Researchers develop single cell genomics technique to reverse engineer developing lungfrom PhysorgMon, 14 Apr 2014, 13:20:59 UTC